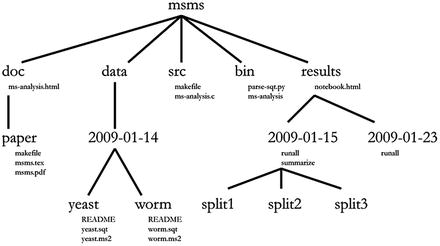
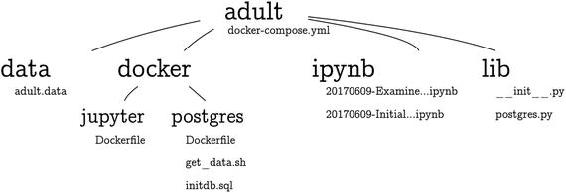
Joshua Cook [Joshua Cook] - Docker for Data Science: Building Scalable and Extensible Data Infrastructure Around the Jupyter Notebook Server
Here you can read online Joshua Cook [Joshua Cook] - Docker for Data Science: Building Scalable and Extensible Data Infrastructure Around the Jupyter Notebook Server full text of the book (entire story) in english for free. Download pdf and epub, get meaning, cover and reviews about this ebook. year: 2017, publisher: Apress, genre: Computer. Description of the work, (preface) as well as reviews are available. Best literature library LitArk.com created for fans of good reading and offers a wide selection of genres:
Romance novel
Science fiction
Adventure
Detective
Science
History
Home and family
Prose
Art
Politics
Computer
Non-fiction
Religion
Business
Children
Humor
Choose a favorite category and find really read worthwhile books. Enjoy immersion in the world of imagination, feel the emotions of the characters or learn something new for yourself, make an fascinating discovery.
- Book:Docker for Data Science: Building Scalable and Extensible Data Infrastructure Around the Jupyter Notebook Server
- Author:
- Publisher:Apress
- Genre:
- Year:2017
- Rating:5 / 5
- Favourites:Add to favourites
- Your mark:
Docker for Data Science: Building Scalable and Extensible Data Infrastructure Around the Jupyter Notebook Server: summary, description and annotation
We offer to read an annotation, description, summary or preface (depends on what the author of the book "Docker for Data Science: Building Scalable and Extensible Data Infrastructure Around the Jupyter Notebook Server" wrote himself). If you haven't found the necessary information about the book — write in the comments, we will try to find it.
It is not uncommon for a real-world data set to fail to be easily managed. The set may not fit well into access memory or may require prohibitively long processing. These are significant challenges to skilled software engineers and they can render the standard Jupyter system unusable.
As a solution to this problem, Docker for Data Science proposes using Docker. You will learn how to use existing pre-compiled public images created by the major open-source technologiesPython, Jupyter, Postgresas well as using the Dockerfile to extend these images to suit your specific purposes. The Docker-Compose technology is examined and you will learn how it can be used to build a linked system with Python churning data behind the scenes and Jupyter managing these background tasks. Best practices in using existing images are explored as well as developing your own images to deploy state-of-the-art machine learning and optimization algorithms.
What Youll Learn
- Master interactive development using the Jupyter platform
- Run and build Docker containers from scratch and from publicly available open-source images
- Write infrastructure as code using the docker-compose tool and its docker-compose.yml file type
- Deploy a multi-service data science application across a cloud-based system
Who This Book Is For
Data scientists, machine learning engineers, artificial intelligence researchers, Kagglers, and software developers
Joshua Cook [Joshua Cook]: author's other books
Who wrote Docker for Data Science: Building Scalable and Extensible Data Infrastructure Around the Jupyter Notebook Server? Find out the surname, the name of the author of the book and a list of all author's works by series.

![Joshua Cook [Joshua Cook] Docker for Data Science: Building Scalable and Extensible Data Infrastructure Around the Jupyter Notebook Server](/uploads/posts/book/119616/thumbs/joshua-cook-joshua-cook-docker-for-data.jpg)




![Dan Toomey [Dan Toomey] - Jupyter for Data Science](/uploads/posts/book/119624/thumbs/dan-toomey-dan-toomey-jupyter-for-data-science.jpg)