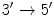This book is meant to serve as an introduction to the new and very exciting field of comparative gene finding. We introduce the field in its current state, and go through the process of constructing a comparative gene finder by breaking it down into its separate building blocks. But before we can dive into the algorithmic details of such a process, we begin by giving a brief introduction to the underlying biological theory. In this chapter we introduce the basic concepts of genetics needed for this book, and define the gene finding problem we have set out to solve. We round off by giving a brief account of the historical developments of approaching the gene finding problem up to where it stands today. In the last section we split the process of building a gene finder into its smaller parts, and the rest of the book is structured in the same manner.
1.1 Some Basic Genetics
Every living organism consists of cells, from just one cell as in the bacterium Escherichia coli ( E. coli ) to many trillions (
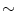
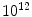
) as in human. Higher organisms also contain considerable amounts of noncellular material, such as bones and water. With a few exceptions each cell contains a complete copy of the genetic material, which is the blueprint that directs all the activities in the organism, and that contains the code for the inheritable traits that are passed from parent to offspring. The genetic material is composed of the chemical substance deoxyribonucleic acid , or DNA for short. single-stranded DNA molecule is a long polymer of small subunits called nucleotides (or bases ). nucleotide consists of a sugar molecule, a phosphoric acid molecule , and one of four nitrogen bases : adenine (A), thymine (T), guanine (G) or cytosine (C), giving rise to the four-letter DNA code {A,T,G,C}. Adenine and guanine belong to the class of purines , while cytosine and thymine belong to the pyrimidines . nitrogen base in purines is slightly larger than that of pyrimidines, consisting of a six-nitrogen and a five-nitrogen ring fused together, while pyrimidines only have a single six-nitrogen ring. In living organisms, DNA usually comes in double-stranded form, where two single-stranded DNA molecules are arranged into a long ladder, coiled to the shape of a double helix . The backbone of the ladder is formed by the sugar and phosphate molecules of the nucleotides, while the rungs of the ladder consist of the nitrogen bases joined in the middle by hydrogen bonds. In this structure, A always binds to T, and G always binds to C, in so-called base pairs (bp), causing the two sides of the ladder, or the two strands in the double helix, to mirror each other.
There are two main types of cells, correspondingly distinguishing between two main types of organisms, namely eukaryotic and prokaryotic cells. Besides the fact that eukaryotic cells are considerably more complex than prokaryotic cells, an important difference is that eukaryotic cells contain a nucleus while prokyarotic cells do not. Most of the genetic material reside in the nucleus in eukaryotes, and is carried on large, physically separate, DNA macromolecules called chromosomes . While the chromosomes in eukaryotes are linear, the DNA in prokaryotes is organized into circular rings. These rings are technically not chromosomes, although many tend to use the term for prokaryotes as well. Each eukaryotic specie has a characteristic number of chromosomes, which for instance is 46 in a typical human cell, 40 in mouse, and only 8 in fruit fly. With typical cells we mean diploid cells, where the chromosomes are organized in pairs . In each pair, one chromosome descends from the mother, and the other from the father. There are two types of chromosome pairs, the autosomes and the sex chromosomes . an autosome pair the two individual chromosomes are of the same length, carry the same inheritable traits, and the number of copies of the chromosomes is the same in both males and females. The sex chromosomes, on the other hand, may have very different characteristics and are also the main indicator of the gender of the organism. The human genome consists of 23 chromosome pairs, including 22 autosomes and one pair of sex chromosomes, X and Y. A female carries the pair XX while males carry the pair XY, where the Y chromosome naturally always comes from the father. The ploidy of an organism signifies the number of copies of the unique set of chromosomes in that organism. Thus, a diploid cell contains two copies of each distinct chromosome (except for the sex chromosomes), whereas a haploid cell bears only one copy of each. Most cells in higher organisms are diploid, but specifically the gametes (the sperm and ova) are haploid. Examples of haploid organisms are fungi, wasps, and ants, and for instance, plants may switch between a haploid and a diploid, or even a polyploid state.
The genetic material on the chromosomes is organized into subunits called the genes of the organism. The genes are subsequences of DNA spread out along the chromosomes, and are intervened by possibly very long stretches of nonfunctional DNA. The genes provide the templates for the proteins and RNA molecules that are responsible for all activity in the organism, and are traditionally defined as the units of inheritance that control the hereditary traits passed on from parent to offspring. The genome of an organism is its complete set of DNA (or RNA for some viruses), including both the genes and the nonfunctional stretches of DNA. The genome sizes vary greatly between organisms; from 600,000 bp in the smallest (known) free-living organism (a bacterium) to some 3 billion bp in human. While the genome is very compact in lower organisms, with very little nonfunctional material, in higher organisms the genes are hidden in a vast sea of junk DNA. In human, for instance, the genes constitute only about 3 % of the human genome. If the rest of the sequence really is junk, or if it has some sort of function, direct or indirect, is still under much debate [].
Fig. 1.1
Each nucleotide consists of a phosphoric acid molecule (P), a sugar ring, and a nitrogen base. The antiparallel strands of the DNA double helix run in opposite directions. The direction is given using the
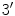
and
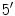
carbon atoms of the sugar rings in the nucleotides. Reading the sequences from left to right, one will have the
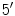
atom to the left of the
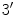
, while in the antiparallel strand the situation is reversed
Although mirroring one another, the chromosome strands run in opposite directions, and are said to be antiparallel . One strand is called the
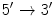
, the forward , or the sense strand , while the other the
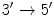










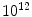 ) as in human. Higher organisms also contain considerable amounts of noncellular material, such as bones and water. With a few exceptions each cell contains a complete copy of the genetic material, which is the blueprint that directs all the activities in the organism, and that contains the code for the inheritable traits that are passed from parent to offspring. The genetic material is composed of the chemical substance deoxyribonucleic acid , or DNA for short. single-stranded DNA molecule is a long polymer of small subunits called nucleotides (or bases ). nucleotide consists of a sugar molecule, a phosphoric acid molecule , and one of four nitrogen bases : adenine (A), thymine (T), guanine (G) or cytosine (C), giving rise to the four-letter DNA code {A,T,G,C}. Adenine and guanine belong to the class of purines , while cytosine and thymine belong to the pyrimidines . nitrogen base in purines is slightly larger than that of pyrimidines, consisting of a six-nitrogen and a five-nitrogen ring fused together, while pyrimidines only have a single six-nitrogen ring. In living organisms, DNA usually comes in double-stranded form, where two single-stranded DNA molecules are arranged into a long ladder, coiled to the shape of a double helix . The backbone of the ladder is formed by the sugar and phosphate molecules of the nucleotides, while the rungs of the ladder consist of the nitrogen bases joined in the middle by hydrogen bonds. In this structure, A always binds to T, and G always binds to C, in so-called base pairs (bp), causing the two sides of the ladder, or the two strands in the double helix, to mirror each other.
) as in human. Higher organisms also contain considerable amounts of noncellular material, such as bones and water. With a few exceptions each cell contains a complete copy of the genetic material, which is the blueprint that directs all the activities in the organism, and that contains the code for the inheritable traits that are passed from parent to offspring. The genetic material is composed of the chemical substance deoxyribonucleic acid , or DNA for short. single-stranded DNA molecule is a long polymer of small subunits called nucleotides (or bases ). nucleotide consists of a sugar molecule, a phosphoric acid molecule , and one of four nitrogen bases : adenine (A), thymine (T), guanine (G) or cytosine (C), giving rise to the four-letter DNA code {A,T,G,C}. Adenine and guanine belong to the class of purines , while cytosine and thymine belong to the pyrimidines . nitrogen base in purines is slightly larger than that of pyrimidines, consisting of a six-nitrogen and a five-nitrogen ring fused together, while pyrimidines only have a single six-nitrogen ring. In living organisms, DNA usually comes in double-stranded form, where two single-stranded DNA molecules are arranged into a long ladder, coiled to the shape of a double helix . The backbone of the ladder is formed by the sugar and phosphate molecules of the nucleotides, while the rungs of the ladder consist of the nitrogen bases joined in the middle by hydrogen bonds. In this structure, A always binds to T, and G always binds to C, in so-called base pairs (bp), causing the two sides of the ladder, or the two strands in the double helix, to mirror each other.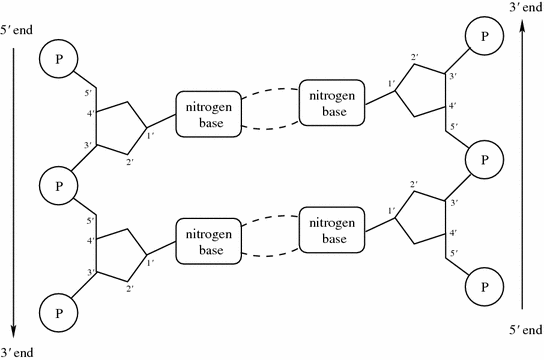
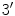 and
and 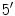 carbon atoms of the sugar rings in the nucleotides. Reading the sequences from left to right, one will have the
carbon atoms of the sugar rings in the nucleotides. Reading the sequences from left to right, one will have the 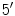 atom to the left of the
atom to the left of the 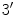 , while in the antiparallel strand the situation is reversed
, while in the antiparallel strand the situation is reversed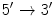 , the forward , or the sense strand , while the other the
, the forward , or the sense strand , while the other the