Christoph Bleidorn - Phylogenomics
Here you can read online Christoph Bleidorn - Phylogenomics full text of the book (entire story) in english for free. Download pdf and epub, get meaning, cover and reviews about this ebook. year: 0, publisher: Springer International Publishing, Cham, genre: Detective and thriller. Description of the work, (preface) as well as reviews are available. Best literature library LitArk.com created for fans of good reading and offers a wide selection of genres:
Romance novel
Science fiction
Adventure
Detective
Science
History
Home and family
Prose
Art
Politics
Computer
Non-fiction
Religion
Business
Children
Humor
Choose a favorite category and find really read worthwhile books. Enjoy immersion in the world of imagination, feel the emotions of the characters or learn something new for yourself, make an fascinating discovery.
- Book:Phylogenomics
- Author:
- Publisher:Springer International Publishing, Cham
- Genre:
- Year:0
- Rating:4 / 5
- Favourites:Add to favourites
- Your mark:
- 80
- 1
- 2
- 3
- 4
- 5
Phylogenomics: summary, description and annotation
We offer to read an annotation, description, summary or preface (depends on what the author of the book "Phylogenomics" wrote himself). If you haven't found the necessary information about the book — write in the comments, we will try to find it.
Phylogenomics — read online for free the complete book (whole text) full work
Below is the text of the book, divided by pages. System saving the place of the last page read, allows you to conveniently read the book "Phylogenomics" online for free, without having to search again every time where you left off. Put a bookmark, and you can go to the page where you finished reading at any time.
Font size:
Interval:
Bookmark:
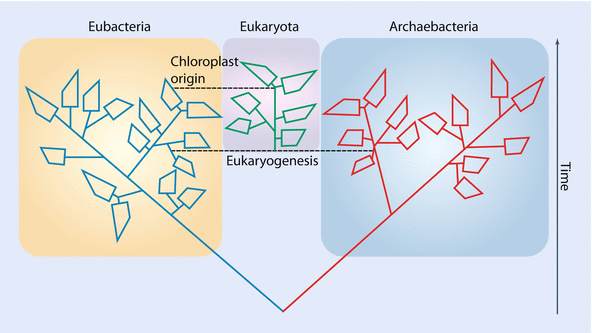
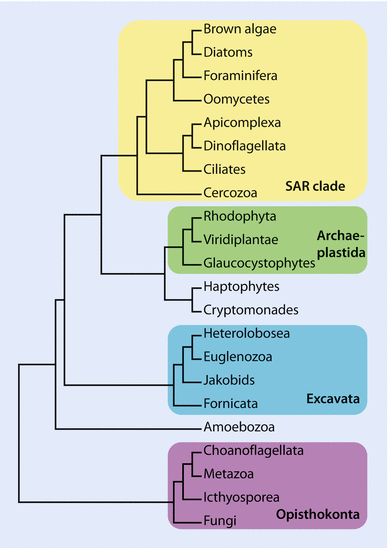
- Life on earth can be largely classified into Bacteria, Archaea and Eukaryota.
- Eukaryotes likely arose by symbiogenic origin due to the fusion of an archaean with a bacterium.
- Bacteria and Archaea have compact genomes with uninterrupted genes, contained by a single, circular DNA molecule, located in the nucleoid.
- Eukaryote genomes are linearly organized into separate chromosomes, located in the nucleus, and contain genes interrupted by introns.
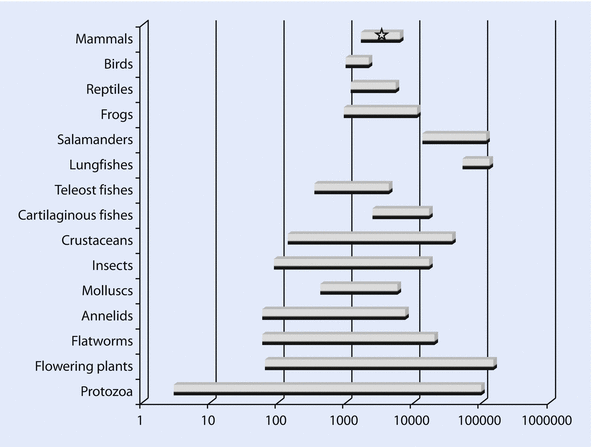
- Eukaryotes bear substantially larger genomes than archaeans and bacteria, but within eukaryotes there is no correlation between complexity and genome size.
- The human genome is around 3.3 Gb in size, but protein-coding genes and other functional DNA only make up a small proportion (<10%), whereas transposable elements are dominating (>44%).
- High-throughput sequencing of ancient human DNA allowed the reconstruction of archaic human genomes and led to the discovery of a hitherto unknown lineage, called Denisovan.
Font size:
Interval:
Bookmark:
Similar books «Phylogenomics»
Look at similar books to Phylogenomics. We have selected literature similar in name and meaning in the hope of providing readers with more options to find new, interesting, not yet read works.
Discussion, reviews of the book Phylogenomics and just readers' own opinions. Leave your comments, write what you think about the work, its meaning or the main characters. Specify what exactly you liked and what you didn't like, and why you think so.












