Springer Science+Business Media LLC 2017
Paul Wood (ed.) Lipidomics Neuromethods 10.1007/978-1-4939-6946-3_1
1. Introduction and Overview of Lipidomic Strategies
William J. Griffiths 1 and Yuqin Wang 1
(1)
Swansea University Medical School, Singleton Park, Swansea, SA2 8PP, UK
William J. Griffiths
Email:
Abstract
While analysis of the genome or proteome can be predictive of the fate of an organism, its metabolome is informative of the outcome of events on an organism. The metabolome can thus be regarded as closer to the actual phenotype than either the genome or proteome. Lipids represent a major component of the metabolome and their hydrophobic and amphipathic nature dictates their separate analysis from more water-soluble metabolites; hence, the discipline of lipidomics has emerged. In this short chapter, we will highlight the different strategies in lipidomics which have now reached maturity, i.e., shotgun and chromatography-based mass spectrometry approaches. We will discuss some of the newer technologies coming to the forefront e.g., the use of derivatization chemistry, and comment on exciting developments now being made in the lipidomic field, e.g., surface analysis and lipid imaging. Finally, we will comment on some of the dangers encountered using an omics approach in biochemical analysis.
Introduction
Lipidomics can be defined as the quantitative identification of all lipids in an organism, tissue, fluid, or cell type. Lipids themselves are defined as hydrophobic or amphipathic small molecules that may originate entirely or in part by carbanion-based condensations of thioesters (e.g., fatty acyls) and/or by carbocation-based condensations of isoprene units (e.g., prenols, sterols) [].
MS gives molecular weight information via measurement of the mass-to-charge ratio ( m/z ) of ionized species and many modern MS instruments are capable of providing mass accuracy at a level of 0.0010.002 m/z . This degree of mass accuracy allows m/z searches against, e.g., the Lipid Maps database of compounds ].
MS not only provides information for lipid identification, it can also be used for lipid quantification. Ideally, quantification is performed with the use of isotope-labeled standards added during sample preparation, or if these are not available via structurally related analogues. Avanti Polar Lipids http://avantilipids.com/ provide a large number of high-quality authentic standards imperative for accurate quantification.
There are essentially two strategies in lipidomics analysis. There is the global shotgun approach developed by Han and colleagues in the USA [].
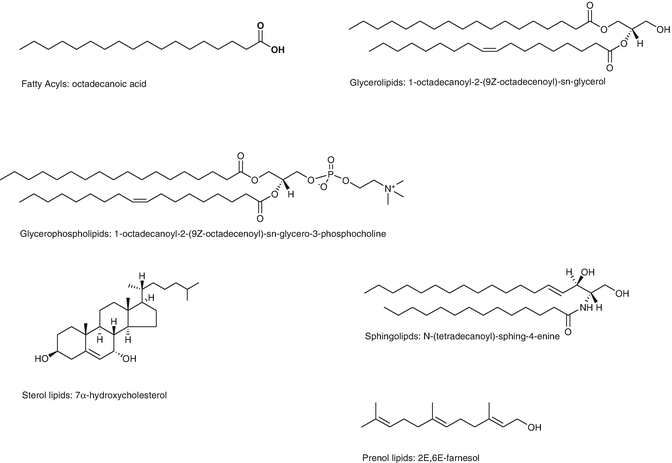
Fig. 1
Six classes of lipids commonly analyzed in lipidomics studies
Shotgun Lipidomics
The term shotgun lipidomics was coined by Han and Gross and the methodology was excellently reviewed in their 2005 publication []. The scanning protocols used depend on the mass spectrometer available, but the goal is always to identify and quantify as many lipid molecular species as possible.
2.1 Intra-Source Separation and MDMS
Han and Gross performed their initial studies using tandem quadrupole instruments and designed their workflow to take advantage of the performance characteristics of these instruments [].
2.2 Top-Down Lipidomics
Shevchenko and colleagues in Dresden have developed a shotgun lipidomics approach, called top-down lipidomics, exploiting the attributes of high-resolution mass spectrometers [].
2.3 Shotgun Steroidomics
While it is fair to say that in the era of ESI the dominant methodologies have been those developed by the groups, and colleagues, of Han and Schevchenko, in the preceding era dominated by fast atom bombardment (FAB)-MS and liquid secondary ionization mass spectrometry (LSIMS) similar methods were used for metabolite profiling, particularly for steroids and bile acids. FAB-MS and/or LSIMS have been extensively used to profile steroids and bile acids in urine [].
2.4 LC-Shotgun Lipidomics
While the classic shotgun lipidomics experiment involves direct infusion of a lipid extract, this can be modified by the up-stream inclusion of LC column offering some separation before ionization []. The inclusion of an LC step does have its disadvantages. Throughput is reduced as the LC separation will have a defined run time, cross-contamination between injections is always a worry as is overloading of the column and distorting quantitative measurements. It should also be remembered the cruder the sample the more likely the column will become blocked or its performance deteriorate.
Targeted Lipidomics
An alternative to the shotgun lipidomic approach is provided by targeted lipidomics, where each class of lipid is analyzed one by one, exploiting extraction and MS procedures specifically designed for the target class of analyte. This approach is the one adopted by the Lipid Maps consortium in the US ].
3.1 Targeted Lipidomics Exploiting Derivatization
The idea of using derivatization chemistry to target specific functional groups and enhance the analysis of compounds possessing these groups has been exploited in MS for decades []. They targeted the O -alkenyl-ether double bond within plasmenyl ethers by derivatization with iodine and methanol. The derivatized molecules show characteristic mass shifts and give informative fragment ions upon MS/MS. They also derivatized amine-containing plasmalogens with 13C1- S , S -dimethylthiobutanoylhydroxysuccinimide ester (13C1-DMBNHS) reagent prior to iodine and methanol addition. Again, derivatization provides a characteristic mass shift and informative fragment ions are forthcoming upon MS/MS.
We have developed a derivatization strategy for the analysis of sterols and steroids . Inspired by the use of the Girard hydrazine reagents by Shackleton et al. to enhance the ESI-MS ion-current for steroids possessing a carbonyl group [].
While most chemical derivatization reactions are performed prior to MS analysis. Derivatization can also be exploited within the mass spectrometer itself. Blanksby, Mitchell, and colleagues in Australia have developed a derivatization method they call OzID in which selected precursor ions are subjected to gas phase reaction with ozone in a linear ion-trap []. The resulting CID/OzID spectra are particularly valuable for determining sn- position and carbon double bond positions in glycerophospholipids.
Surface Analysis and Mass Spectrometry Imaging
Modern mass spectrometry imaging (MSI) has been pioneered by Richard Caprioli and colleagues in Vanderbilt University [].
Surface analysis can also be achieved using ESI via the technique desorption electrospray ionization (DESI) developed by Graham Cooks and colleagues in Purdue []. DESI generally provides lower lateral resolution than MALDI-MSI but avoids the necessity for matrix application.
Conclusions
Lipidomics has progressed over the last decade to a mature technology []. There are still major challenges ahead, e.g., in the localization of lipids to defined spatial regions, the observation of their temporal changes, and the analysis of lipids that may be minor in most situations and only become abundant after appropriate stimulation. There are also dangers for lipid analysis using an omics approach. The availability of metabolite and lipid databases has led to a careless use of the term identification. To an analytical chemist a compound is only identified when its chemical and physical properties exactly match those of an authentic standard. When only a single property, e.g., exact mass, is matched to a structure in a database, the term annotate is more appropriate. Care must also be taken when performing quantification. The gold standard for quantification is isotope dilution MS where an isotope labeled standard is added during analyte extraction. This may not always be possible in which case quantification is best performed against structural analogues. Finally, with respect to quantification, the analyst should be aware of the precision of their method by analysis of repeated quality control samples before claiming changes in analyte concentrations are a consequence of a biological pertebation. With these caveats in mind the future is bright.
















![Wood - Cia Rose Series Box Set [Books 1-3]](/uploads/posts/book/141420/thumbs/wood-cia-rose-series-box-set-books-1-3.jpg)

