Katarzyna J. Bandyra
Ben F. Luisi
INTRODUCTION
It may seem surprising that in almost all known life-forms, information-encoding transcripts are actively annihilated. Although at first glance this seems to be a potential waste of resources and loss of information, the anticipated advantages of restricting transcript lifetimes include fast response rates and a capacity to rapidly redirect gene expression pathways. In this way, destroying individual transcripts in a modulated manner might effectively enhance the collective information capacity of the living system. Escherichia coli has proven to be a useful model system to study such processes, and nearly 45 years ago, a hypothetical endoribonuclease was proposed by Apirion as the key missing factor that might account for the observed degradation patterns of mRNA in that bacterium. At the time this hypothesis was formulated, transcript decay in E. coli was best described as a series of endonucleolytic cleavages and subsequent fragment scavenging by 3 exonucleases ().
In the ensuing decades following the discovery of RNase E, more evidence and deeper insights have been gained into the function and importance of the enzyme in RNA metabolism. The data corroborate the numerous roles played by the RNase, including the initiation of turnover for many mRNA species ().
It is important to note that RNase E is not the sole RNase that can initiate turnover in E. coli , as others can catalyze the initial cleavage of mRNAs, including RNase G, RNase P, the double-strand-specific RNase III, and RNases from the toxin/antitoxin families (), implicating a unique and dominating role.
The access of RNase E and other RNases to substrates can be modulated by RNA-binding proteins (). These local structures can be induced or remodeled by base-pairing interactions formed in cis or trans , or by other binding proteins and the unwinding/remodeling activity of helicases. The actions of all these factors modulate substrate access.
In the degradation pathway of mRNA for E. coli , the initial cleavage of a transcript by RNase E is followed closely by exonucleolytic degradation of the products by PNPase (polynucleotide phosphorylase), RNase II, or RNase R ().
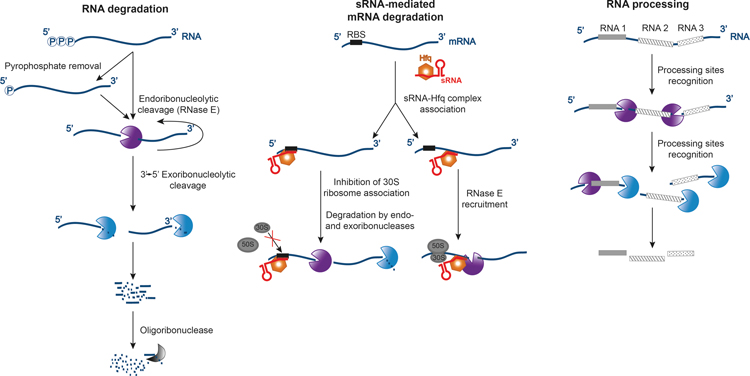
Figure 1: RNase-dependent processes in bacteria. RNases play crucial roles in efficient removal of defective or unnecessary RNAs, regulation of gene expression by sRNAs, and processing of various types of RNAs. (Left) RNA degradation is initiated by endoribonucleolytic cleavage, which can be preceded by pyrophosphate removal from the primary transcript. The majority of degradation initiation events are RNase E dependent. The initial cleavage generates monophosphorylated RNA fragments that can either boost subsequent RNase E cleavage or become substrates for cellular exoribonucleases. Fragments resulting from exoribonucleolytic degradation are further converted to nucleotides by oligoribonuclease. (Middle) When RNA degradation is mediated by sRNA, sRNA-chaperone complexes (such as sRNA-Hfq) can recognize a complementary sequence near the translation initiation region and prevent ribosome association on the transcript (left branch). Naked mRNA is rapidly scavenged by endo- and exoribonucleases. The sRNA-Hfq complex can also bind within the coding region of mRNA, recruiting RNase E and promoting transcript decay (right branch). (Right) In the case of substrates for processing, the order of RNA processing can be defined by the structure of precursors and the specificity of the RNases. The processing can form a cascade of interdependent events where some target sites are being revealed only upon specific initial cleavage. RNA, dark blue; endoribonucleases, purple; exoribonucleases, light blue; sRNA, red; ribosomes, gray ovals; Hfq, orange.











