Introduction
In fluorescence in situ hybridization (FISH) a probe that contains a modified nucleotide in several positions binds to a complimentary nucleotide sequence. The modified nucleotide is then detected by immunofluorescence. FISH can be used (a) to detect the chromosomal locations of genes, BACs, cosmids, and repetitive sequences or to identify whole chromosomes (DNA FISH) and (b) to detect transcripts both at transcription sites in the nucleus and in the cytoplasm (RNA FISH). Classically DNA FISH has been employed to identify which chromosome carries a particular gene or BAC sequence in a metaphase spread, to identify chromosomal deletions or translocations, or to identify chromosomes from the two parents in hybrids (Fig. ].
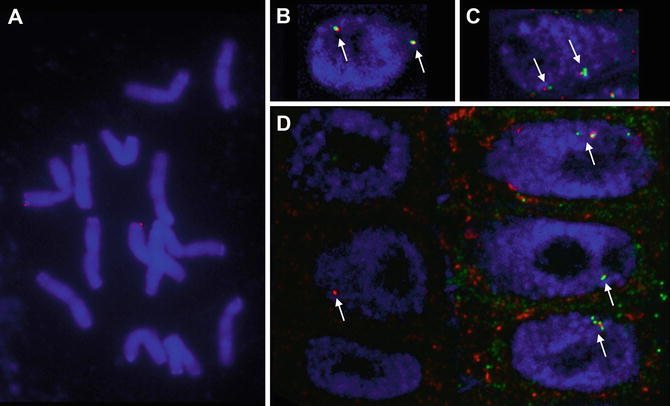
Fig. 1
Examples of DNA and RNA FISH in oat. ( a ) Metaphase plate showing the position of the Sad1 gene ( red ). ( b and c ) DNA FISH on G1 nuclei ( blue ) in oat roots reveals the positions of the Sad1 ( red ) and Sad2 ( green ) gene ( arrows ). Both genes are highly expressed in the epidermis ( c ), while Sad2 is not expressed in the subepidermis at all and Sad1 to a lesser degree ( b ). As a result the 80 kb region spanning the two genes is decondensed in the epidermis and condensed in the subepidermis. ( d ) RNA FISH reveals nascent transcripts of Sad1 ( red ) and Sad2 ( green ) in nuclei of the subepidermis on the left and the epidermis on the right ( arrows ). The cytoplasmic localizations of the two transcripts in epidermis cells do not overlap and there is no Sad2 transcript in the subepidermis. ( b d ) Overlay of several optical sections
DNA probes are used for DNA FISH and RNA probes for RNA FISH (Subheadings ). During hybridization denatured DNA probes are supposed to bind to both strands of the denatured target genomic sequences. In RNA FISH, the probe (an antisense transcript) forms a double strand with the sense target transcript in the cell. In principle, RNA probes could also be used for DNA FISH as the stability of double-stranded nucleotide sequences increases from DNADNA to DNARNA to RNARNA. However, single-stranded RNA probes are susceptible to degradation by RNAses, which are ubiquitous, so care must be taken to work in as clean an environment as possible and DNA in situ hybridization experiments commonly include an RNase step to remove transcripts, which would increase background hybridization. DNA probes are usually prepared by nick translation. Nick translation mixes or kits are commercially available and their use is recommended. RNA probes are prepared by in vitro transcription, for which enzyme mixes are available and their yield is significantly greater than that of nick translation reactions. Genomic sequences down to 5 kb can be detected by DNA FISH and transcript probes should be at least 500 bp in length.
Subheading describes the immunodetection.
Tissue sections were also prepared from root tips of Avena strigosa seedlings. Other tissues might need modifications to the protocol where indicated. For both DNA and RNA FISH, tissue is harvested and fixed in freshly prepared 4 % formaldehyde, which is superior to commercial formaldehyde solutions. It is then embedded in wax in a tissue processing machine and sectioned on a rotary microtome (Subheading ).
For DNA FISH, tissue sections are dewaxed and treated with cell wall degrading enzymes to facilitate probe and antibody penetration (Subheading describes the immunodetection.
For RNA FISH, tissue sections are also dewaxed and treated with cell wall degrading enzymes to facilitate probe and antibody penetration (Subheading describes the immunodetection.
For imaging of metaphase spreads and semithin tissue sections, a widefield microscope with a good-quality cooled CCD or an sCMOS camera is preferable to a scanning confocal microscope because the sensitivity of the former exceeds that of even modern GaAsP or hybrid detectors, which is important for weak signals from small or low-abundancy probes. When using a widefield microscope, deconvolution, which removes out of focus blur, will greatly improve the quality of image stacks from tissue sections and should always be used. If deconvolution software is not available, then a confocal microscope might give better results. Metaphase spreads do not need deconvolution.