1.1 An Example R Session
Here is a simple R session.
> help(sleep)
> x1 <- sleep$extra[sleep$group == 1]
> x2 <- sleep$extra[sleep$group == 2]
> t.test(x1, x2)
> sleep[c(1:3, 11:13), ]
> with(sleep, t.test(extra[group == 1],
+ extra[group == 2]))
> q()
The help() command prints documentation for the requested topic. The sleep dataset is a built-in dataset in R. It comes from William Sealey Gossets article under the pseudonym Student (1908). It contains the effects of two drugs, measured as the extra hours of sleep as compared to controls. The vectors x1 and x2 are assigned the values of the extra hours of sleep in drugs 1 and 2, respectively. (a less than sign followed by a minus sign, <- , represents assignment) Two equal signs, == , represent the logical equal operator. The t.test(x1, x2) carries out an independent sample t -test of the sleep time between the two groups. The same analysis can be done using with(sleep, t.test(extra[group == 1], extra[group == 2])) . sleep[c(1:3, 11:13), ] prints observations 1 through 3 and 11 through 13. To exit the R program, type q() . Typing q without the parentheses prints out the contents of the function to quit R. Most functions are visible to the user in this way. The advantage of using built-in datasets is that they have already been imported. The next example describes how to import data from a text file.
The sleep data can be entered into a text file, the variable names on the first row, and the variables are separated by spaces.
extra group ID
0.7 1 1
-1.6 1 2
-0.2 1 3
-1.2 1 4
-0.1 1 5
3.4 1 6
3.7 1 7
0.8 1 8
0.0 1 9
2.0 1 10
1.9 2 1
0.8 2 2
1.1 2 3
0.1 2 4
-0.1 2 5
4.4 2 6
5.5 2 7
1.6 2 8
4.6 2 9
3.4 2 10
Suppose the data entries are saved in a file named t1.dat in the directory C:\\Documents and Settings\\usr1\\My Documents , then this command imports the data and assigns it a name called sleep.df .
> sleep.df <- data.frame(read.table(file =
+"C:/Documents and Settings/usr1/My Documents/t1.dat",
+header = TRUE))
On a Windows platform, the double back slashes () in a path name can be replaced with one forward slash ( / ). On Unix/Linux and Mac OS, one forward slash works fine. The read.table() function reads the data in file . It uses the first line of the raw data file ( header = TRUE ) to assign variable names to the three columns. Blank spaces in the raw data file are ignored. The data.frame() function converts the imported data into a data frame. The sleep.df data is now available for analysis (type objects() to see it). The example above shows some of the unique features of R. Most data analytic tasks in R are done through functions, and functions have parameters such as the options of file and header in the read.table() function. Functions can be nested, the output of one function can be fed directly into another. Some other basic R features are covered in the next section. These features make R flexible but more challenging to learn for beginners.
Some things are more difficult with Respecially if you are used to using menus. With R, it helps to have a list of commands in front of you. There are lists in the on-line help and in the index of An introduction to R by the R Core Development Team, and in the reference cards listed in http://finzi.psych.upenn.edu/ .
Some things turn out to be easier in R. Although there are no menus, the on-line help files are very easy to use, and quite complete. The elegance of the language helps too, particularly those tasks involving the manipulation of data. The purpose of this book is to reduce the difficulty of the things that are more difficult at first. Next we will go over a few basic concepts in R. The remainder of this chapter covers a few examples on how to take advantage of Rs strengths.
1.2 A Few Useful Concepts and Commands
1.2.1 Concepts
In R, most commands are functions. The command is written as the name of the function, followed by parentheses, with the arguments (inputs) of the function in parentheses, separated by commas when there is more than one, e.g., plot(swiss) to plot a pairwise scatterplot of the swiss data. When there is no argument, the parentheses are still needed, e.g., q() to exit the program. A function is said to return its output when the output is printed or when we can set a variable equal to the output. For example, sqrt(4) returns (prints) 2 on the screen; and if we say v1 <- sqrt(4) , v1 is set equal to the output of the function, or 2.
Some basic concepts in R are surprising to beginners. For example, the square of
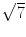
is not 7.
> 7 == sqrt(7)^2
[1] FALSE
That is because floating point arithmetic is not exact.
> options(digits = 22)
> sqrt(7)^2
[1] 7.000000000000000888178
A solution is to compare all.equal(sqrt(7)^2, 7) .
In this book, we generally use names such as x1 or file1 , that is, names containing both letters and a digit, to indicate variable names that the user makes up. Really, these can be of any form. We use the number simply to clarify the distinction between a made up name and a key word with a predetermined meaning in R. R is case sensitive; for example, X and x can stand for different things. We generally use upper-case data objects like X , Y , and M to represent matrices or arrays; and lower-case objects to represent vectors. Although most commands are functions with the arguments in parentheses, some arguments require specification of a key word with an equal sign and a value for that key word, such as source("myfile1.R", echo = T) , which means read in myfile1.R and echo the commands on the screen. It helps to add spaces between input parameters, so that the extra spaces in echo = T make it easier to read than echo=T . But that is not necessary. Key words can be abbreviated (e.g., e = T ). In addition to the idea of a function, R has objects and modes. Objects are anything that you can give a name. There are many different classes of objects. The main classes of interest here are vector , matrix , factor , list , and data frame . The mode of an object tells what kind of things are in it. The main modes of interest here are logical , numeric , and character .
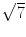 is not 7.
is not 7.








