To explain how one-dimensional patterns choreograph the world we live in, we have no choice but to take a close look at how sequences function at the molecular level. As Howard Pattee says, we should first test our basic concepts at the cellular level where we know more about how it works.
At one extreme we find the sequences of speech and writing which we all know very well, so well, in fact, that we may even think we have good intuitions about how they do what they do. At the other extreme we find the sequences of DNA, RNA, and proteins in the cell. Here scientists have given us an ever-more-detailed understanding of how sequences go about their business. But unless you are a molecular biologist who keeps up with the literature, chances are good that this is a subject you never studied, or that you have studied but largely forgotten, or that you studied so long ago that the field has moved on.
I wish this book could have been written without discussing the details of how cells work but, as you will see, that was not possible. Fortunately, you dont need to study all 912 pages of The Molecular Biology of the Gene to get up to speed; just read this brief chapter now and refer back to it as necessary. I have made every effort to pare it down to only those topics that come up in the book.
Some molecules can bond to one another and assemble themselves end-to-end into long chains, hundreds, thousands, millions of molecules long. To get the idea, imagine an extremely long freight train in which each boxcar is coupled to each of its neighbors. These chains are polymers, and their individual boxcars are monomers. All of our common plastics are made from these chains. Polymers put the poly in polyethylene, polypropylene, polystyrene, and PVC (polyvinyl chloride). These everyday polymer chains are just the same monomer over and over again ad infinitum, like a train in which all of the boxcars are identical. I call these chains rather than sequences because they are patternless; any chain 100 monomers long looks just like any other chain 100 monomers long.
Bio polymers are a different story because in the living world it is common for chains to be made from more than one kind of monomer, not just boxcars, but flatbeds, tankers, hoppers, etc. As a result, two chains 100 monomers long can exhibit different patterns; these patterned chains can properly be called sequences. The number of possible patterns depends on how many different kinds of monomers are in the alphabet, as well as the length of the chain. It does not take very many monomers or a very long chain to yield a huge number of possibilities.
Three biopolymers are of interest to us: DNA, RNA, and protein. Well start with DNA and RNA, which are close cousins, both made up of nucleic acids, otherwise known as nucleotides, nucleotide bases, or just bases. Each has four different kinds of nucleotide, four different letters in its molecular alphabet. The nucleotides in DNA are abbreviated A, C, G, and T; those in RNA are A, C, G, and U. You can see that DNA and RNA have three bases in common (A, C, G) and each has one that is different, T in DNA and U in RNA.
When we describe sequences of DNA, the order of bases in the tiny molecule specifies the pattern of the abbreviations. For example, GATTACA represents a unique sequence of seven nucleotide bases, just one of many possible arrangements. In fact, there are 16,384 unique ways to arrange the four bases into such a seven-base sequence, which is a lot of diversity. When you stop to consider that a typical gene can be hundreds of bases long, you quickly realize that the number of potential genes is mind-boggling, even with only a four-base alphabet.
For now, lets just say that a gene is a very long sequence of DNA that guides the construction of a single protein. The set of all genes in a cell is called its genome. Even simple cells contain thousands of genes and make thousands of proteins. In most cases when we discuss sequences of DNA or RNA, we pay attention to their one-dimensional patterns and not to their three-dimensional shapes. There are interesting exceptions, however, and we will meet them in due course.
Proteins are the other major group of biopolymers; sometimes they are called polypeptides or peptide chains. Their alphabet comprises 20 different monomers, each a kind of amino acid, sometimes amino acid residues or just residues. As we shall see, proteins are initially constructed as sequences, but they normally fold themselves up into three-dimensional shapes. When we discuss proteins, sometimes we pay attention to their linear pattern and sometimes to their shape, and sometimes to both.
Sequences of DNA and RNA have the important and unusual property of being easy to replicate. This is because their nucleotide bases can bond to other bases in more than one direction, not only horizontally (lengthwise) to form a linear pattern but also vertically, at a right angle to the direction of the sequence. In the usual horizontal direction, each base bonds equally well with any of the four varieties. However, in the vertical direction each will bond with only a single unique partner. This makes faithful replication possible.
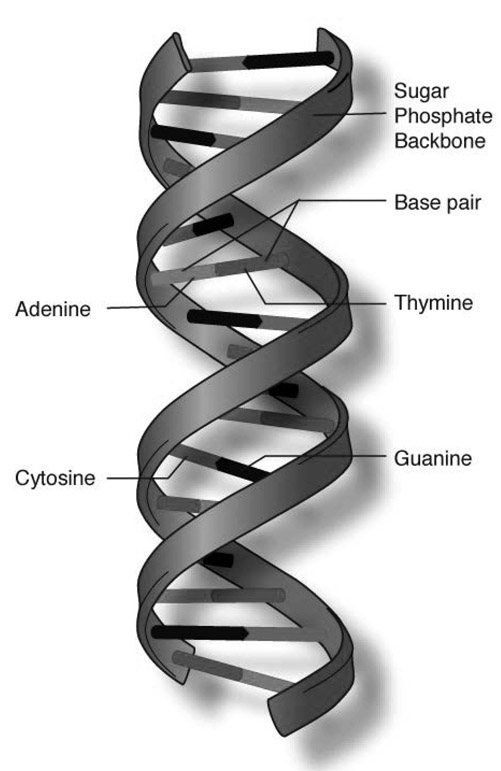
FIGURE A.1 This stylized drawing shows the double helix of the DNA molecule, with nucleotide bases (A, T, C, G) matched with their complements.
Source: National Human Genome Research Institute (www.genome.gov/genetics-glossary/Double-Helix)
For example, to form a horizontal sequence, an A will bond with another A, or a T, C, or G. However, in the vertical direction A bonds exclusively with T, and C only with G, and vice versa. The same applies to RNA, except that A bonds with U rather than T. Any two complementary bases bonded together like this are called a base pair.
Imagine a seven-monomer DNA sequence like GATTACA. If each nucleotide pairs vertically with its partner (C to G, A to T, etc.), then the result is the complementary sequence CTAATGT. This process is called template matching or base-pairing or sometimes Watson-Crick pairing, after the discoverers of DNAs structure. The original sequence provides a template on which its complement is constructed. Scale this up to the billions of base pairs in the genome and this is how DNA replicates during cell division.
If you look at a DNA molecule in a cell, you will notice it exists in double-stranded form, two complementary sequences bonded to one another. When not busy with something else, each nucleotide base remains naturally bonded to its complement. This yields the famous double helix structure of DNA.
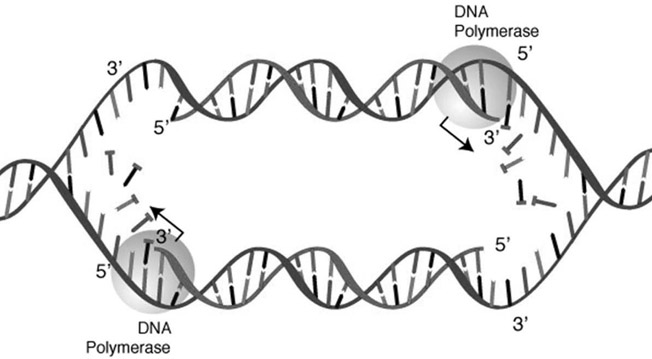
FIGURE A.2 A high-level view of DNA replication. The double helix is unwound and unzipped, and DNA polymerase constructs complementary sequences on the two exposed strands.
Source: National Human Genome Research Institute (www.genome.gov/Pages/Careers/EducationalPrograms/ShortCourse/2016ShortCourse/2016-08-03ShortCourseGeneticsandGenomicsPrimer_BW.pdf)
It also adds to another necessary step when it is time to replicate. Since every nucleotide is already bonded to its complement, that bond must be broken to allow a new strand to be formed. The double helix must be unwound and unzipped, and two new complementary sequences built upon both single-stranded templates. The job of unzipping the DNA and building the complementary sequences falls to a team of molecules centered on the enzyme













