Gerhard Steger - DNAzymes : Methods and Protocols
Here you can read online Gerhard Steger - DNAzymes : Methods and Protocols full text of the book (entire story) in english for free. Download pdf and epub, get meaning, cover and reviews about this ebook. publisher: Springer US, genre: Home and family. Description of the work, (preface) as well as reviews are available. Best literature library LitArk.com created for fans of good reading and offers a wide selection of genres:
Romance novel
Science fiction
Adventure
Detective
Science
History
Home and family
Prose
Art
Politics
Computer
Non-fiction
Religion
Business
Children
Humor
Choose a favorite category and find really read worthwhile books. Enjoy immersion in the world of imagination, feel the emotions of the characters or learn something new for yourself, make an fascinating discovery.
- Book:DNAzymes : Methods and Protocols
- Author:
- Publisher:Springer US
- Genre:
- Rating:4 / 5
- Favourites:Add to favourites
- Your mark:
- 80
- 1
- 2
- 3
- 4
- 5
DNAzymes : Methods and Protocols: summary, description and annotation
We offer to read an annotation, description, summary or preface (depends on what the author of the book "DNAzymes : Methods and Protocols" wrote himself). If you haven't found the necessary information about the book — write in the comments, we will try to find it.
DNAzymes : Methods and Protocols — read online for free the complete book (whole text) full work
Below is the text of the book, divided by pages. System saving the place of the last page read, allows you to conveniently read the book "DNAzymes : Methods and Protocols" online for free, without having to search again every time where you left off. Put a bookmark, and you can go to the page where you finished reading at any time.
Font size:
Interval:
Bookmark:

For further volumes: http://www.springer.com/series/7651
For over 35 years, biological scientists have come to rely on the research protocols and methodologies in the critically acclaimed Methods in Molecular Biology series. The series was the first to introduce the step-by-step protocols approach that has become the standard in all biomedical protocol publishing. Each protocol is provided in readily-reproducible step-by-step fashion, opening with an introductory overview, a list of the materials and reagents needed to complete the experiment, and followed by a detailed procedure that is supported with a helpful notes section offering tips and tricks of the trade as well as troubleshooting advice. These hallmark features were introduced by series editor Dr. John Walker and constitute the key ingredient in each and every volume of the Methods in Molecular Biology series. Tested and trusted, comprehensive and reliable, all protocols from the series are indexed in PubMed.
This Humana imprint is published by the registered company Springer Science+Business Media, LLC part of Springer Nature.
The registered company address is: 1 New York Plaza, New York, NY 10004, U.S.A.
Ribozymesenzymes consisting of only RNAwere well established in 1990: Thomas Czech and coworkers had established the self-splicing of Tetrahymena group I intron [1, 2], Sidney Altman and coworkers had described the ribozyme nature of RNAse P cleaving off a precursor of tRNA [3, 4], George Bruening and coworkers had detected the self-cleavage activity of satellite RNAs [5], and Robert Symons and coworkers had described the self-cleavage activity of viroids from family Avsunviroidae [6]. At this time, most scientists expected that such ribozyme activity would be restricted to RNA because DNA misses several features that were thought to be essential for catalytic activity: DNA occurs mostly not single-stranded and thus rarely forms complex tertiary structures, and DNA lacks the 2-OH group of RNA. Thus, the first publication on a DNAzymeDNA with enzymatic activityby Ronald Breaker and Gerald Joyce [7] in 1994 came as a big surprise. Today, a broad variety of DNAzymes with different catalytic functions are known [8], including RNA cleavage [7], peptide modification [9, 10], phosphorylation [11], thymine dimer photoreversion [12], peroxidation [13], and DNA ligation [14]. Schematic representations of four selected DNAzymes are shown in Fig..
The DNAzyme described by Breaker and Joyce in 1994 and most of the many DNAzymes described afterwards were found by in vitro selection starting from a random pool of DNA molecules, called systematic evolution of ligands by exponential enrichment (SELEX) [16]. The principle of SELEX to obtain molecule(s) with a desired function is based on the assumption that a three-dimensional structure, which is required for function, can be formed by many different single-stranded sequences. In theory, a random pool of sequences with 15 up to 50 nucleotides in length and the four different nucleotides comprises 415 1.2 1018 up to 450 1.3 1030 different sequences, covering all possible structures within the limits of the sequence length; in practice this number of random sequences is limited to about 1014 to 1015 sequences due to additionally required constant sequence parts, for example for amplification, hybridization, and fixation. One key step in the selection process is the separation of nucleic acids with the desired functionality from those which lack this property and therefore should be excluded from the pool in the next selection round. The other key step is amplification of the relatively low amount of recovered nucleic acids with the desired functionality after each selection round by polymerase chain reaction (PCR) [17, 18], which requires fixed primer binding sites at the ends of the random sequence part.
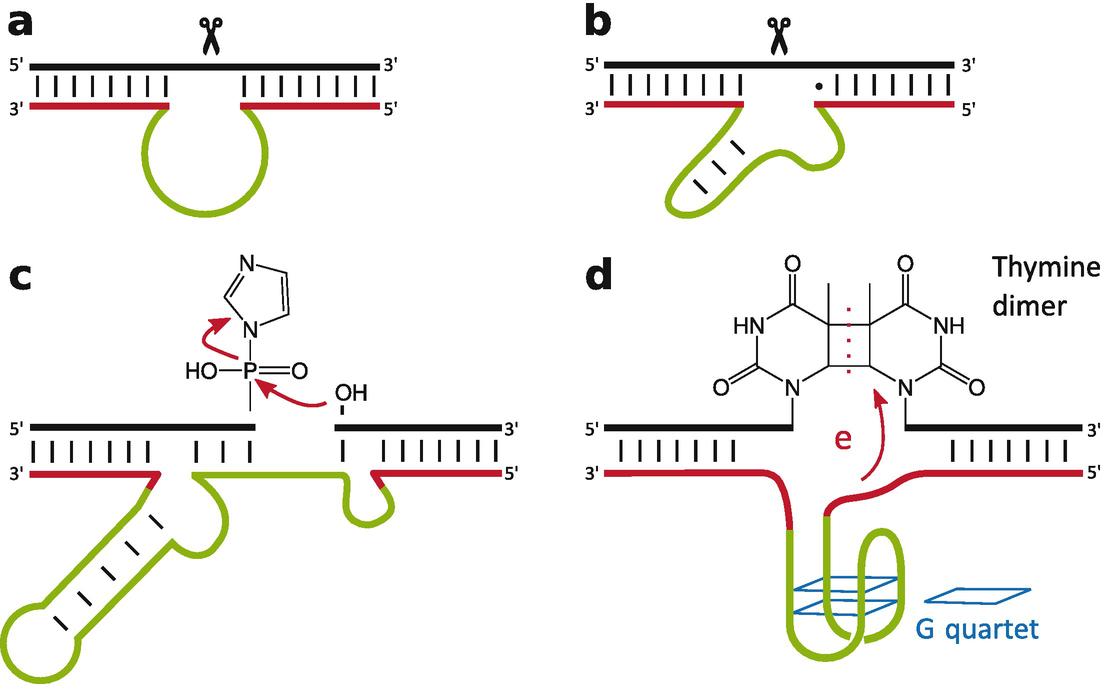
Schematic representation of different DNAzymes: The RNA-cleaving DNAzymes (a) 10-23 and (b) 8-17, (c) the DNA-ligating DNAzyme E47, and (d) the UV1C DNAzyme with photolyase activity. Catalytically important sequences are represented in green. The binding arms, which can vary in their sequence, are shown in red. The target sequences are shown in black. (Figure based on [15])
For example, the SELEX procedure (Fig. shows a schematic representation of the in vitro selection process for RNA-cleaving DNAzymes.
Similar procedures were used to produce the nowadays great variety of DNAzymes cleaving RNA in the presence of various metal ions and at various conditions [22, 23]. If one aims at other catalytic properties of DNAzymes than RNA cleavage, different immobilization and PCR methods are required; examples of such methods are given in Chapters 1 and 2 by Li et al. and Yang et al. SELEX requires the recovery or modification of single-stranded oligonucleotide sequence pools from mixed template libraries. Chapter 3 by Szokoli et al. provides two protocols for the PCR-based production of ssDNA molecules from low amounts of starting material.
Font size:
Interval:
Bookmark:
Similar books «DNAzymes : Methods and Protocols»
Look at similar books to DNAzymes : Methods and Protocols. We have selected literature similar in name and meaning in the hope of providing readers with more options to find new, interesting, not yet read works.
Discussion, reviews of the book DNAzymes : Methods and Protocols and just readers' own opinions. Leave your comments, write what you think about the work, its meaning or the main characters. Specify what exactly you liked and what you didn't like, and why you think so.













