1.1.1 Graphs, Nodes, and Arcs
A graph G =( V , A ) consists of a nonempty set V of nodes or vertices and a finite (but possibly empty) set A of pairs of vertices called arcs , links , or edges .
Each arc a =( u , v ) can be defined either as an ordered or an unordered pair of nodes, which are said to be connected by and incident on the arc and to be adjacent to each other. Since they are adjacent, u and v are also said to be neighbors . If ( u , v ) is an ordered pair, u is said to be the tail of the arc and v the head ; then the arc is said to be directed from u to v and is usually represented with an arrowhead in v ( u v ). It is also said that the arc leaves or is outgoing for u and that it enters or is incoming for v . If ( u , v ) is unordered, u and v are simply said to be incident on the arc without any further distinction. In this case, they are commonly referred to as undirected arcs or edges , denoted with e E and represented with a simple line ( u v ).
The characterization of arcs as directed or undirected induces an equivalent characterization of the graphs themselves, which are said to be directed graphs (denoted with G =( V , A )) if all arcs are directed, undirected graphs (denoted with G =( V , E )) if all arcs are undirected, and partially directed or mixed graphs (denoted with G =( V , A , E )) if they contain both directed and undirected arcs.
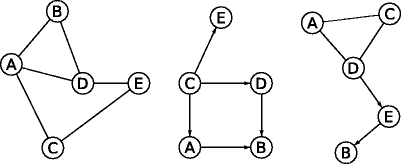
Examples of directed , undirected , and mixed partially directed graphs are shown in Fig.:
The node set is V ={ A,B,C,D,E} and the edge set is E ={ (AB), (AC), (AD), (BD), (CE), (DE) }.
Arcs are undirected, so, i.e., AB and BA are equivalent and identify the same edge.
Likewise, A is connected to B, B is connected to A, and A and B are adjacent.
Fig. 1.1
An undirected graph ( left ), a directed graph ( center ) and a partially directed graph ( right )
For the directed graph, Fig :
The node set is V ={ A,B,C,D,E} and the graph is characterized by an arc set A ={ (A B), (CA), (DB), (CD), (C E)} instead of an edge set E .
Arcs are directed, so, i.e., AB and BA identify different arcs. For instance, AB A while BA A . Under the additional constraint of acyclicity, it is not possible for both arcs to be present in the graph because there can be at most one arc between each pair of nodes.
Also, A and B are adjacent, as there is an arc (AB) from A to B. AB is an outgoing arc for A (the tail), an incoming arc for B (the head), and an incident arc for both A and B.
On the other hand, the partially directed graph, Fig., is characterized by the combination of an edge set E ={ (A C), (AD), (C D)} and an arc set A ={ (D E), (E B)}.
An undirected graph can always be constructed from a directed or partially directed one by substituting all the directed arcs with undirected ones; such a graph is called the skeleton or the underlying undirected graph of the original graph.
1.1.2 The Structure of a Graph
The pattern with which the arcs appear in a graph is referred to as either the structure of the graph or the configuration of the arcs. In the context of this book it is assumed that the vertices u and v incident on each arc are distinct and that there is at most one arc between them so that ( u , v ) uniquely identifies an arc. This definition also implicitly excludes presence of a loop that can occur when u = v .
The simplest structure is an empty graph , i.e., a graph with no arcs. On the other end of the spectrum are saturated graphs , in which each node is connected to every other node. Real-world graphical abstractions usually fall between these two extremes and can be either sparse or dense . While the distinction between these two classes of graphs is rather vague, a graph is usually considered sparse if
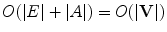
.
The structure of a graph can reveal interesting statistical properties. Some of the most important ones deal with paths . Paths are essentially sequences of arcs or edges connecting two nodes, called end-vertices or end-nodes . Paths are denoted with the sequence of vertices ( v 1, v 2,, v n ) incident on those arcs. The arcs connecting the vertices v 1, v 2,, v n are assumed to be unique, so that a path passes through each arc only once. In directed graphs it is also assumed that all the arcs in a path follow the same direction, and we say that a path leads fromv 1 (i.e., the tail of the first arc in the path) tov n (i.e., the head of the last arc in the path). In undirected and mixed graphs (and in general when referring to a graph regardless which class it belongs to), arcs in a path can point in either direction or be undirected. Paths in which v 1= v n are called cycles and are treated with particular care in Bayesian network theory.
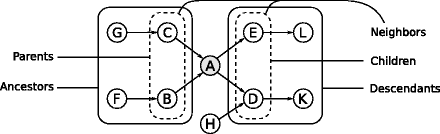
The structure of a directed graph defines a partial ordering of the nodes if the graph is acyclic , that is, if it does not contain any cycle or loop. This ordering is called an acyclic or topological ordering and is induced by the direction of the arcs. It is defined as follows: if a node v i precedes v j , there can be no arc from v j to v i . According to this definition the first nodes are the root nodes , which have no incoming arcs, and the last ones are the leaf nodes , which have at least one incoming arc but no outgoing ones. Furthermore, if there is a path leading from v i to v j , v i precedes v j in the sequence of the ordered nodes. In this case v i is called an ancestor of v j and v j is called a descendant of v i . If the path is composed by a single arc, by analogy x i is a parent of v j and v j is a child of v i .
Consider, for instance, node A in the directed acyclic graph shown in Fig.. Its neighborhood is the union of the parents and children; adjacent nodes necessarily fall into one of these two categories. Its parents are also ancestors, as they necessarily precede A in the topological ordering. Likewise, children are also descendants.










 .
.