1.1 ProteinDNA Interactions
Deoxyribo-nucleic acids (DNA), which carry genetic information, and proteins which execute and regulate life process are two important biomolecules in any living organisms. The dynamical interactions between proteins and DNA play a central role in many biological processes such as DNA replication, transcription, recombination, gene regulation, repair, and even the packaging of DNA into chromosomes. In order to interact with DNA and perform their designated functions, proteins must first recognize and bind specifically to their target site on DNA. The study of the dynamics of proteinDNA interactions aimed at unraveling the mechanisms of binding-site recognition is a fascinating research field in biophysics and the central focus of this dissertation.
There are two classes of DNA-binding proteins: (1) proteins that bind to DNA in a sequence-independent manner (nonspecific binding), e.g., wrapping of DNA along the histone cores in nucleosome; (2) proteins that bind to DNA in a sequence-dependent manner by recognizing specific sites in DNA (site-specific binding), e.g., binding of transcription factors. Although in general, site-specific proteins are highly selective and bind to their target DNA sites with thousand- to million-fold higher affinities compared to random DNA sequences, some others, for example, some DNA damage recognition proteins, recognize a broad range of target sites with only 10100-fold higher affinity than nontarget sites [].
Two puzzles have particularly intrigued researchers in this field. The first seeks to understand how site-specific DNA-binding proteins efficiently search for their binding sites in DNA among a sea of nonspecific sites. The second related puzzle is how this protein actually recognizes its target site on genomic DNA. This question is less well understood and is the main topic of this thesis. The recognition problem can be separated from the search problem by focusing on short DNA oligomers that contain a target sequence for a specific DNA-binding protein and by investigating the dynamics of the interactions that lead to the formation of the specific complex.
When proteins bind to DNA, they often distort the DNA at the binding site by bending, kinking, or twisting the DNA. As an example, genomic DNA is smoothly but severely bent when it is wrapped around the histone octamers to form nucleosomes for chromosome packaging []. These examples illustrate that DNA deformability and concerted rearrangements in the bound protein to accommodate the deformed DNA is an essential aspect of nearly all biological phenomena involving proteinDNA interactions and likely play an essential role in the mechanism by which proteins recognize their target sites in genomic DNA.
While X-ray crystallography and NMR have been extraordinary in elucidating the three-dimensional structures of proteinDNA complexes and providing some insights into the recognition mechanisms, these structural studies dont reveal the time scales or the order in which these conformational changes occur, thus leaving a big gap in our understanding of the energetics of these interactions. Studying the dynamics of these conformational changes is important because the time scales on which these structural changes occur are inherently related to the biological function.
1.2 Sequence-Dependent DNA Deformability and Its Role in Target Recognition
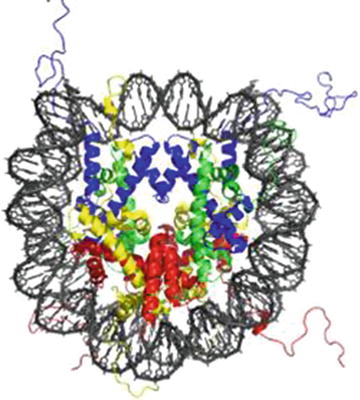
It is well recognized that DNA has remarkable flexibility and can undergo considerable distortions in order to adopt many different conformations when bound to a protein. As discussed above, a classic example of distorted DNA is found in the building blocks of the chromosome: the nucleosomal core particle (NCP), in which ~147 bp of DNA are wrapped approximately twice around the histone core (Fig. ).
Fig. 1.1
The crystal structure of DNA wrapped around the histone core to form nucleosome. Nucleosome consists of four core histones H2A ( yellow ), H2B ( red ), H3 ( blue ), and H4 ( green ). The view is from the top through the super helical axis. PDB:1AOI []
High-resolution crystal structures of the NCP, first obtained by Richmond, Lugar, and coworkers in 1997, showed that the DNA is not uniformly bent within the nucleosome but rather it is periodically kinked to adopt the donut-like surface of the histone octamer core [].
These and several other studies showed that the persistence length of DNA is not a constant for a given set of conditions but in fact depends on the sequence context. For example, alternating AT polymers is 2030% more flexible than mixed sequence DNA [].
These conclusions are consistent with the kinked DNA seen in NCP crystal structures, as discussed above, and imply that the sequence-dependent structural and mechanical properties of a DNA segment can strongly influence protein binding by reducing the energy cost of adopting a particular conformation suitable for protein binding.
1.2.1 Free Energy Cost for Local Deformation of DNA
The structure and stability of DNA has contributions from both base-stacking energy, which is from hydrophobic and van der Waals interactions between bases, and base-pairing energy, which is from hydrogen bonding between complementary bases. A smaller contribution to the helix stability also comes from phosphatephosphate repulsions on the outside of the helix [].
Fig. 1.2
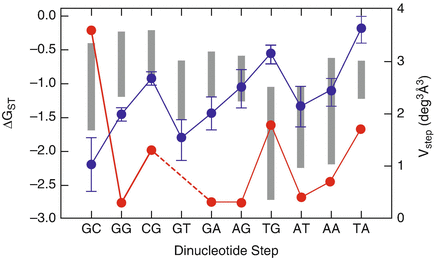
Nearest-neighbor (nn) stacking free energies ( G ST) for the ten different dinucleotide steps of duplex DNA. The nn stacking free-energy parameters, extracted from thermal denaturation experiments on oligonucleotide duplexes, are shown as vertical gray bars . The length of each vertical bar indicates the rage of the stacking parameters from seven independent research groups, obtained under different salt conditions and for varying length of duplex DNA, and unified by SantaLucia. Stacking free energies from electrophoretic mobility measurements ( filled blue circle ) on DNA fragments containing a nick in the sugar-phosphate backbone, between all possible combinations of dinucleotide steps are from Fran-Kamenetskiis data. The volume of thermally accessible conformational space for each dinucleotide step ( filled red circle ), obtained from fluctuation and correlations of base step parameters in DNAprotein crystal complexes. (Figure reproduce from [] with permission)
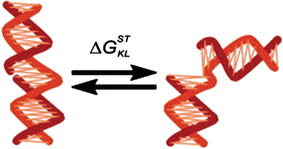
In another study, Frank-Kamenetskii and coworkers used electrophoretic gel mobility measurements of a series of 300 bp long DNA molecules nicked in the sugar phosphate backbone on one of the strands, with the nick sandwiched between all combinations of dinucleotide steps. The nick alters the equilibrium between stacked (straight) and unstacked at the nick (bent) conformations (Fig. ].
Fig. 1.3
Schematic representation of the stacked-to-unstacked conformational transition of the DNA fragment at the nick site described by the energy difference, G STKL (Figure reprinted from [], copyright (2004), with permission from Elsevier