1.1 Tuberculosis in History
Tuberculosis (TB) has caused more deaths through the last 200 years than any other infectious disease, and has been with us since ancient times (Paulson 2013). Evidence of tuberculosis has been found in 9,000 year old mummies. There are conflicting theories of the timing of the emergence of Mycobacterium tuberculosis ( M.tuberculosis ) as a human pathogen with two recent theories proposing 70,000 years ago (Comas et al. 2013) or 6,000 years ago. The later study proposed that seals first transmitted the disease to humans (Bos et al. 2014).
Tuberculosis (TB) is a chronic granulomatous disease caused by the bacterium M. tuberculosis , and more rarely, other species of the Mycobacterium tuberculosis complex including Mycobacterium bovis and Mycobacterium africanum. The term tubercle in the context of consumptive (wasting) disease was first coined by Fransiscus de la Bo (also known as Sylvius of Leyden), a Dutch anatomist in the 17th century. He found tubercles (from: tuberculum, small lump in Latin) in the lungs of most consumptives. Before the discovery of the pathogen in 1882 by Robert Koch, the spectrum of diseases caused by the mycobacteria were known by many names including: consumption, phtisis (from Greek phtinein to waste away), scrofula (swelling of lymphnodes, especially in the neck), Potts disease (tuberculous spondylitis, named after a British orthopedic surgeon Percivall Pott, in the 18th century, but found in Egyptian mummies and art as early as 1000 BC), yaksma (from Sanskrit: gradual destruction) and shaky oncay (Incan), balasa (Hindu: swelling). The European epidemic in the 17th century was known as the white plague (Fig. ).
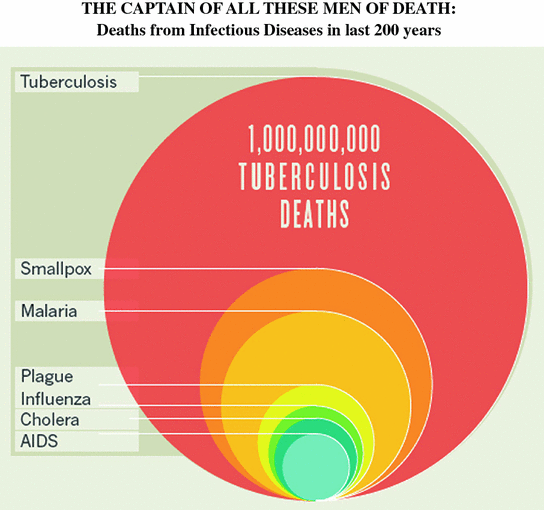
Fig. 1.1
The burden of tuberculosis. From Paulson T. Nature, 2013. Reprinted with permission
1.2 Pathogen
TB is caused by bacteria of the Mycobacterium tuberculosis complex, mostly M.tuberculosis , but rarely also M.canetti, M.microti, M.africanum, and M.bovis (de Jong et al. 2010). Mycobacteria are non-motile, non spore-forming, aerobic, rod-shaped bacteria of 24 m in length and possess a unique lipid-rich cell wall which gives the acid-fast property by which they are known (acid-fast bacilli, or AFBs) and renders them resistant to many disinfectants and antibiotics. They can be divided into slow growing or rapid growing species (Image ).
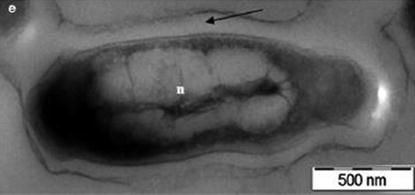
Image 1.1
Transmission electron microscope image of Mycobacterium tuberculosis . The Black arrow indicates the thick myolic acid layer. The n . indicates the nucleide (from Srinivasan et al., Arch Microbiol, 2014, reprinted with permission)
M. tuberculosis is slow-growing, non-pigmented and appears as cream coloured breadcrumbs on culture, often also described as rough, tough and buff (Collins 1997) (Image . The only other major human pathogen of the mycobacteria genus is M. leprae, which causes leprosy and is not discussed further (White and Franco-Paredes 2015).
Image 1.2
Mycobacterium tuberculosis colonies on solid Lowenstein Jensen medium ( courtesy of Dr. Dang Thi Minh Ha)
The whole genome of M. tuberculosis (laboratory strain H37Rv) was sequenced in 1998 (Cole et al. 1998). Subsequent sequencing of clinical strains from around the world has illuminated pathogen diversity, evolution and spread (Comas et al. 2013). Six major geographic lineages of M. tuberculosis have been identified: the Euro-American, Indo-Oceanic, East-Asian (including Beijing strains), West-African 1 and 2, and East-African-Indian. Many studies have attempted to identify lineage-specific differences in clinical virulence and/or transmissibility, but results have been conflicting. These different findings may be the result of differences in the particular strains used for comparison, variation in host genetics, environmental influences or different study methodologies.
Some strains (e.g. Beijing and Haarlem strains) have been associated with increased drug resistance. This may result from intrinsic factors such as increased genetic mutation rates, intrinsic drug tolerance, lower fitness cost associated with resistance-conferring mutations (Ford et al. 2013), or from environmental factors that facilitated its emergence and spread. Current typing methods such as spoligotyping, IS6110 restriction fragment length polymorphism (RFLP) and variable number tandem repeat (VNTR) have value for outbreak investigations and studies of population transmission, but do not offer any information to guide treatment. Advances in the speed and cost of whole genome sequencing will soon supersede other typing techniques and would be far more informative, facilitating detailed transmission mapping and proving information on likely drug-resistance to guide clinical management (Anderson et al. 2014; Comas et al. 2013; Barry et al. 2012; Borrell and Gagneux 2009; Borrell et al. 2013; Cohen et al. 2011; Coll et al. 2013; Steiner et al. 2014).
1.3 Epidemiology
Although TB is often thought of as a historical disease in the developed world, this is not the case. Globally in 2012 there were an estimated 8.6 million new cases of active TB and 1.3 million deaths; therefore there is one new TB case every 4 s and more than two TB deaths every minute. Twenty-two high-burden countries account for 80 % of all TB cases, with India and China alone contributing almost 40 % (26 and 12 % respectively). The TB incidence per 100,000 population varies dramatically, from less than 10 per 100,000 in developed countries such as Japan, the United States, Western Europe and Australia, to rates exceeding 1000 per 100,000 in South Africa and Swaziland (WHO 2014). Overall, it is estimated that just 64 % of incident TB cases were notified to National TB Programmes in 2013 (WHO 2014).