Springer International Publishing AG 2017
Amanda L. Garner (ed.) RNA Therapeutics Topics in Medicinal Chemistry
Computational Tools for Design of Selective Small Molecules Targeting RNA: From Small Molecule Microarrays to Chemical Similarity Searching
Matthew G. Costales 1, Jessica L. Childs-Disney 1 and Matthew D. Disney 1
(1)
Departments of Chemistry and Neuroscience, The Scripps Research Institute, 130 Scripps Way, Jupiter, FL 33458, USA
Abstract
RNA is an important drug target, yet few lead compounds elicit their effects by acting on RNA outside of the bacterial ribosome. Herein, we describe various synergistic strategies to identify small molecules that target RNA and how computational approaches can be utilized for lead optimization. In particular, we describe the development of small molecule microarray approaches applied towards RNA and its application to identify small molecule binders and for the facile study of antibiotic resistance mechanisms for known or novel lead antibacterials. Additionally, a microarray-based library-versus-library screen, which probes millions of combinations, is described that identifies RNA motif binding partners preferred by small molecules. Lead compounds can be designed by searching for these privileged interactions in a disease-causing RNA. Computational chemistry can be used to optimize these compounds. For example, lead compounds that target the r(CCUG) repeats expansions that cause myotonic dystrophy type 2 (DM2) were lead optimized by using structure-based design. Specifically, the compounds were developed to allow an in situ click chemistry approach in which a disease-affected cell synthesizes its own drug on-site by using the disease-causing biomolecule as a cellular catalyst. In another lead optimization strategy, chemical similarity searching was employed to lead optimize small molecules that target the r(CUG) repeat expansion that causes myotonic dystrophy type 1 (DM1). These studies allowed for the identification of an in vivo active small molecule that targets r(CUG) and improves disease-associated defects. As more studies are completed to understand the role of RNA in disease biology, the number of potential RNA targets will increase. In order to leverage these important investigations to develop compounds that target RNA, approaches that allow one to identify and optimize small molecules for selectivity and potency must be carefully considered.
Introduction
RNA has been long viewed as an important drug target in both bacterial and human systems. The earliest examples of small molecules that target RNA include antibiotics binding to various sites in the bacterial ribosome [].
The use of small molecules to target human RNAs has been limited, but there have been a few important examples that include affecting pre-mRNA splicing by binding to an mRNA [].
In this review, we describe various approaches to identify lead small molecules that target RNA. An emphasis is placed on various bead- and microarray-based screening approaches to identify and study lead RNA-targeting small molecules []. In the coming years, these approaches are likely to advance a paradigm in which cell permeable small molecules can be quickly designed to target a disease-causing RNA.
RNA Targeting and the Transcriptome: Structured RNAs Abound!
Since the early days in RNA biology, it was well known that RNAs, including ribosomal (rRNA) and transfer (tRNAs) RNAs, adopt a folded structure [].
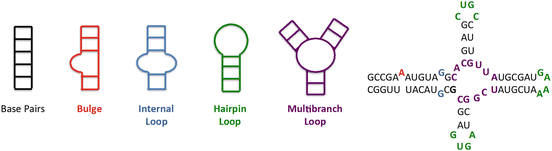
Fig. 1
Secondary structural elements formed when single stranded RNA folds on itself to minimize its free energy. RNA can form canonical base pairs (GC, AU, and GU pairs) or noncanonically paired regions including bulges, internal loops (both symmetric and asymmetric), hairpin loops, and multibranch loops. The different elements are color coded in the secondary structure on the right
RNA folding has been studied ever since, including investigations on phylogenetic comparison of rRNA to identify the kingdoms of life and even to define the new ones [].
A renaissance in RNA structural biology was put forward by X-ray crystallography [].
One major advance emanating from studies on the ribosome included defining the binding pockets of various antibiotics [].
RNA Targets: Binding to Isolated Fragments Mimics Binding to the Biologically Active Structure
In many of the above examples, small molecule screens were completed to identify and study small molecule binding to isolated regions of a biologically active RNA target. The works of Wong []. Chemical mapping of ribosomal particles bound to paromomycin showed that dynamic switching affected decoding of tRNAs and explained the antibacterial activity of these small molecules.
Studies by Pilch, Hermann, and Tor advanced these studies in isolated systems and showed that the ability of aminoglycosides to affect dynamics of this site was a better predictor of antibacterial efficacy than simple binding [].
Small Molecule Microarrays for Studying Ligand Binding
The Schreiber group developed the small molecule microarray approach by arraying small molecules onto surfaces in a spatially defined manner []. Compounds on the array bound a labeled protein target, indicated by signal at specific positions. Thus, this approach provided a facile method to score compounds for binding to targets.
Inspired by these studies, Disney and coworkers developed a small molecule microarray approach to profile the binding of small molecules to RNA targets. These studies were used to profile aminoglycosides for binding to isolated A-sites from both the human and bacterial ribosome and identify small molecules that were not substrates for resistance-causing enzymes []. The use of the microarray method to study binding of RNA targets has been extended by us and others and is gaining widespread use.
Two-Dimensional Combinatorial Screening: Facile Profiling of Libraries of RNA Motifs for Binding to Libraries of Small Molecules
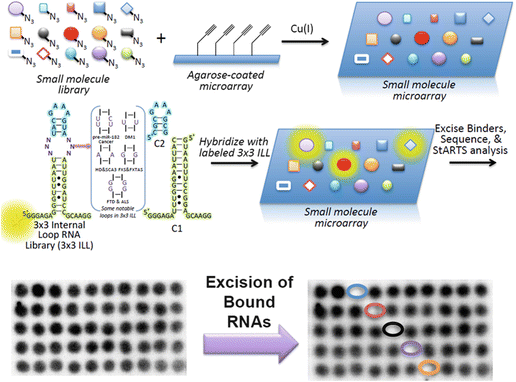
Given the developments of profiling a single or few RNA targets for binding to array-immobilized ligands, we sought to develop a novel method to profile large-scale molecular recognition events via microarray (Fig. ].
Fig. 2
Schematic of Two- D imensional C ombinatorial S creening (2DCS), a library-vs.-library screening approach, to identify the RNA motifs preferred by small molecules []. Briefly, small molecules are site-specifically immobilized onto a functionalized agarose-coated microarray, for example, by using click chemistry. The arrays are hybridized with a labeled RNA library that displays discrete secondary structural elements such as internal loops that are likely to be found in biological RNAs. Hybridization is completed under conditions of high stringency using oligonucleotides that mimic regions that are common to all members of the RNA library (C1 and C2). Bound RNAs, as visualized by deposition of signal on the array surface, are excised from the array, amplified, and sequenced. The RNA-small molecule pairs are then deposited into a database that can be used as a facile lead compound identification strategy (Inforna)