Cytologic specimen may contains less amount of target cells compared with formalin-fixed paraffin-embeded (FFPE) surgical specimen. However, cytologic specimens, especially those obtained through fine needle aspiration, are often more suitable for molecular assays due to the high quality nucleic acids by non-formalin fixation and less fragmented genome.
Selection of Molecular Methods
In addition to considering clinical utility as initial step, several factors should be considered before conducting validation testing of a molecular assay. They include: (1) types of genetic alteration, such as amplification, mutation, indels, and gene fusion; (2) clinical sensitivity and specificity; (3) accuracy, precision and detection of low limit; (4) simplicity, associated with shorter turn-around time and lower cost; (5) availability of tissue type, such as fresh tissue, FFPE or cytologic specimen; (6) clinical volume and cost effective issue.
Common genetic alterations in neoplasm include point mutation, indels, gene fusion, amplification, aneuploidy/polysomy and abnormal methylation. Commonly used molecular assays in clinical lab are polymer chain reaction (PCR), reverse transcriptional PCR (RT-PCR), Florescence in situ hybridization (FISH) and conventional (Sanger) DNA sequencing. Recently new highthrough put molecular technologies, such as DNA/RNA microarray, Sequenoms MassARRAY system and next generation sequencing (NGS) have been introduced and increasingly used in clinical laboratories. In addition, conventional cytogenetic lab is employing more and more new molecular technology, such as FISH and microarray comparative genomic hybridization Testing (array-CGH).
PCR-based assays are suitable for detection of point mutation, small indels, gene fusions (RT-PCR), amplification, and methylation. PCR product (amplicon) is also the first step in harvesting targeted DNA fragment for performing DNA sequencing. FISH assays can be used for detection of gene amplification, indels, gene break-apart (surrogate test for gene fusion), and aneuploidy. Sequenoms MassARRAY and next generation sequencing (NGS) are powerful technologies and can be used to detect almost all types of genetic alterations. Table ).
Table 1.1
Selection of molecular techniques for detection of genetic alterations
Molecular targets | Cytogenetic analysis (metaphase) | PCR (DNA) | RT-PCR (RNA) | FISH | DNA microarray | RNA microarray | Sangers DNA sequencer | Sequenom MassARRAY | Next generation DNA sequencer |
|---|
DNA |
Deletion/insertion | X | X | X | X | X | X | X |
Amplification | X | X | X | X | X | X |
Point mutation/SNP | X | X | X | X | X |
Gene fusion (translocation) | X | X | X | X | X | X | X | X |
Aneuploidy/polysomy | X | X | X | X | X |
Hypermethylation | X | X | X |
Genome | X | X | X | X | X |
RNA |
mRNA expression | X | X | X | X | X |
Gene fusion (translocation) | X | X | X | X | X | X |
miRNA/siRNA | X | X | X | X |
Transcriptome | X | X | X | X | X |
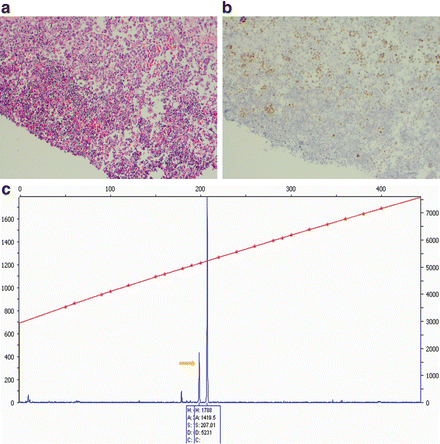
Fig. 1.1
PCR assay for EGFR mutation on cytologic cell block of plural fluid. ( a ) section of cell block of plural fluid with lung adenocarcinoma and inflammatory cells, H&E stain; ( b ) immunostain for TTF-1 highlights the cells of lung adenocarcinoma, making it easier to be isolated by microdissection under microscopy; ( c ) PCR product of EGFR exon 19 by capillary electrophoreses (Genetic Analyzer 310), showing a 12 bp deletion ( arrow )